Difference between revisions of "Protein Community"
AngelHerraez (talk | contribs) (loading with filter) |
AngelHerraez (talk | contribs) (Nice display of membrane proteins from OPM) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 211: | Line 211: | ||
which accelerates rotation (the thickness disappears while manipulating with the mouse) | which accelerates rotation (the thickness disappears while manipulating with the mouse) | ||
| − | + | This command is equivalent: | |
set cartoonFancy on | set cartoonFancy on | ||
| + | |||
| + | See also [[Recycling_Corner#Improved_lighting]] | ||
== Loading part of the model == | == Loading part of the model == | ||
| Line 218: | Line 220: | ||
See documentation ([{{ScriptingDoc}}#loadgeneraloptions 1], [{{ScriptingDoc}}#k549 2]) and [[Load/Filter|practical examples]]. | See documentation ([{{ScriptingDoc}}#loadgeneraloptions 1], [{{ScriptingDoc}}#k549 2]) and [[Load/Filter|practical examples]]. | ||
| + | |||
| + | |||
| + | == Nice display of membrane proteins == | ||
| + | The [https://opm.phar.umich.edu/proteins/ Orientation of Proteins in Membranes] (University of Michigan, Lomize Group) website holds a database with the (approximate) positions of proteins embedded in membranes. The two membrane (lipid bilayer) surfaces are displayed as a set of dummy atoms added to the pdb file, red (O) for the outer side and blue (N) for the inner side. | ||
| + | [[File:OPM renderings.png]] | ||
| + | |||
| + | The following Jmol script locates those dummy atoms, removes them and inserts in their place the respective circular planes, making a cylinder to represent the portion of membrane where the protein is embedded. | ||
| + | You can see the script at work at [https://biomodel.uah.es/model2/prot-membr/get_OPM.htm this page]. | ||
| + | |||
| + | <pre> | ||
| + | /* Code by Angel Herráez */ | ||
| + | load "https://opm-assets.storage.googleapis.com/pdb/6xmt.pdb"; | ||
| + | rotate X -90; /* +Z upwards, outer side */ | ||
| + | define m~out [DUM].O; define m~in [DUM].N; | ||
| + | if ( {m~out}.size>0 ) { | ||
| + | diam~mem = 2.2 * {m~out}.x.max; | ||
| + | z~out = {m~out}.xyz; | ||
| + | if ( {m~in}.size>0 ) { | ||
| + | z~in = {m~in}.xyz; | ||
| + | } else { | ||
| + | z~in = - z~out; | ||
| + | } | ||
| + | } else if ( {m~in}.size>0 ) { | ||
| + | diam~mem = 2.2 * {m~in}.x.max; | ||
| + | z~in = {m~in}.xyz; | ||
| + | z~out = - z~in; | ||
| + | } else { | ||
| + | diam~mem = 52; | ||
| + | z~out = 14.9; | ||
| + | z~in = -14.9; | ||
| + | }; | ||
| + | draw surf~mem diameter @diam~mem color translucent navajoWhite cylinder @z~in @z~out fill; | ||
| + | draw surf~out scale @diam~mem color orangeRed arc @z~out @z~in; | ||
| + | draw surf~in scale @diam~mem color royalBlue arc @z~in @z~out; | ||
| + | delete [DUM]; | ||
| + | select protein; cartoon only; color chain; | ||
| + | </pre> | ||
Latest revision as of 19:52, 10 February 2023
- Community & users
- Communities: Crystal · Protein · Quantum Chemistry · Solid State · Folding@home
- List of users & their pages
This subcommunity is for Jmol users that use Jmol for display and analysis of protein structures. Please add on this page your favorite scripts etc. for displaying proteins in Jmol.
- Angel Herraez page
- Eric Martz: FirstGlance in Jmol is a simple tool for exploring the structure of any on-line macromolecular model. Proteopedia.Org is a Jmol-based wiki with easy Scene-Authoring Tools for creating and sharing customized molecular scenes without learning Jmol language.
Contents
Adjusting speed and quality
Responsiveness to user interaction and quality of the rendering may be adjusted using several options or parameters in JmolScript. See Quality and speed
Molecular surfaces
documentation and testing page
Secondary structure
Starting with versions 12.0.18 and 12.1.15, Jmol implements the DSSP algorithm for determination of secondary structure in proteins. This is accompanied by some changes in the defaults and some new commands.
Starting with Jmol 14.6 the new DSSP 2.0 algorithm is used. This change only affects helices that have bulges, indicating them more appropriately as pi helices rather than alpha helices.
- Rob van der Kant, Gert Vriend (2014) Int. J. Mol. Sci. 15: 7841-7864; doi:10.3390/ijms15057841
- Implemented at http://www.cmbi.ru.nl/dssp.html
- To use DSSP 1.0 instead, you may use
calculate structure DSSP 1.0,show DSSP 1.0,load ... filter "DSSP1"
When reading files
- If the
 pdb or
pdb or  mmcif file contains HELIX/SHEET/TURN information: that information is respected in all Jmol versions. In new Jmol versions,
mmcif file contains HELIX/SHEET/TURN information: that information is respected in all Jmol versions. In new Jmol versions,  pdb and
pdb and  mmcif readers also read helix types alpha, pi, and 310, and color them slightly differently (see colors below).
mmcif readers also read helix types alpha, pi, and 310, and color them slightly differently (see colors below). - If the
 pdb or
pdb or  mmcif file des not contain HELIX/SHEET/TURN information:
mmcif file des not contain HELIX/SHEET/TURN information:
- Old versions of Jmol used Ramachandran-angle based calculation.
- New versions of Jmol use DSSP calculation.
- In the case of alpha-carbon-only chains, all versions of Jmol use the method of Levitt and Greer (doi).
- In new versions of Jmol the calculation based on Ramachandran angles is still available (see below).
Forced recalculation of structure
calculate structure
This command overwrites any secondary structure assignment with a new one:
- In old Jmol versions, calculated using the Ramachandran method.
- In new Jmol versions, calculated using the DSSP method.
Note that, intentionally, Jmol turns off all bioshapes when the calculate structure command is given. So, you will usually want to follow it with cartoons on or something of the sort.
Using the command set structure described here you can define the minimum and maximum psi values that would be valid for each structure type.
New commands
calculate structure ramachandranis available for forcing the old method of calculation (it may have some advantages in certain contexts). It can be shortened tocalculate structure rama.calculate hBonds structuredoes DSSP determining hydrogen bonds only.set defaultStructureDSSP falsewill change the defaults of file load andcalculate structureto use the Ramachandran method; by default, it is true and uses the DSSP method.set dsspCalculateHydrogenAlways falsedoes DSSP enforcing the use of backbone amide H atoms present in the file; by default, it is true and does standard DSSP, ignoring all backbone amide H atoms present in the file and using rough approximations instead.show dsspdisplays a report of the DSSP calculation.set debugdisplays a verbose DSSP calculation.save structure s1(for example) saves in memory the current structure assignment with the internal ID 's1' for later restoring.restore structure s1restores the previously saved assignment.
Customization of the Secondary Structure for Regions
As described here, you can can define individual secondary structure elements as well. Select the portion you want to change first. For example,
select 16-31:A; structure HELIX; cartoon on;to define residues 16-31 of chain A as an alpha helix for cartoon style display.
Obviously these adjustments are to be used sparingly as there is often a reason the secondary structure detection methods within Jmol fail to identify the structure and you don't want to make erroneous assignments. Often such adjustments may be useful when the structure assignment information formerly in the file has been removed as result of some processing of the PDB file and the default structure detection results in a secondary structure that does not perfectly match that defined in the published crystal or NMR structure.
The commands:
structure HELIXstructure SHEETstructure TURNstructure helix310<---NEED TO VERIFY WORKS.
Selection and coloring
Atom expressions related to secondary structure:
| by name | by index | ||
|---|---|---|---|
select structure=-1 |
matches non-protein parts (ligands,...) | ||
select structure=0 |
matches unstructured protein (random coil) | ||
select turn |
or | select structure=1 |
matches turns |
select sheet |
or | select structure=2 |
matches beta strands |
select helix |
or | select structure=3 |
includes all 3 types of helices |
select dna |
or | select structure=4 |
matches DNA |
select rna |
or | select structure=5 |
matches RNA |
select carbohydrate |
or | select structure=6 |
matches carbohydrates |
select helix310 |
or | select substructure=7 |
matches 310 helices (DSSP code "G"). Example: 2JC9.pdb |
select helixAlpha |
or | select substructure=8 |
matches alpha helices (DSSP code "H") |
select helixPi |
or | select substructure=9 |
matches pi helices (DSSP code "I"). Example: 2JC9.pdb after calculate structure |
Note: it is safe to use substructure for all selection commands. For ex., select substructure=2 selects beta strands, but select structure=8 selects nothing.
color structurenow applies different shades of color to alpha, 310 and pi helices, in addition to the classic coloring of helices, beta strands, turns and unstructured portions (see colors below).
label %[substructure]labels with the texts: helixalpha, helix310, helixpi, sheet, turn, none
structure helixAlpha....structure helix310....structure helixPi....
Reference section
- W. Kabsch, C. Sander (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22: 2577-2637. doi:10.1002/bip.360221211
- DSSP Home Page: http://swift.cmbi.ru.nl/gv/dssp/
- M. Levitt, J. Greer (1977) Automatic identification of secondary structure in globular proteins. J. Mol. Biol. 114: 181-239. doi:10.1016/0022-2836(77)90207-8
- Codes for structure types:
| PDB code |
SPSS code |
Jmol codes substructure and name | |
|---|---|---|---|
| right-handed alpha (default) | 1 | H | 8 helixalpha █ |
| right-handed omega | 2 | ||
| right-handed pi | 3 | G | 7 helixpi █ |
| right-handed gamma | 4 | ||
| right-handed 310 | 5 | I | 9 helix310 █ |
| left-handed alpha | 6 | ||
| left-handed omega | 7 | ||
| left-handed gamma | 8 | ||
| 27 ribbon/helix | 9 | ||
| polyproline | 10 | ||
| (beta) strand | 0 first 1 parallel -1 antiparallel |
E | 2 sheet █ |
| turn | T | 1 turn █ | |
| isolated beta-bridge residue | B | ||
| bend | S | ||
| none (unstructured protein) | - | 0 none █ | |
| DNA | 4 dna █ | ||
| RNA | 5 rna █ | ||
| carbohydrate | 6 carbohydrate █ | ||
| other* | -1 █ |
- Helix types in PDB format: columns 39-40 of HELIX record, right-justified.
- Strand sense in PDB format: columns 39-40 of SHEET record, right-justified.
- (*) Other = not protein, not nucleic, not carbohydrate.
Pretty cartoons
Apply this script to get cartoons with a better-than-default look (it will take some more time to rotate them around):
set hermiteLevel -4; set ribbonAspectRatio 12;
or
set hermiteLevel 4; set ribbonAspectRatio 12;
which accelerates rotation (the thickness disappears while manipulating with the mouse)
This command is equivalent:
set cartoonFancy on
See also Recycling_Corner#Improved_lighting
Loading part of the model
The use of load with the filter option allows that Jmol will only read some atoms from the file. This is useful e.g. to avoid the solvent molecules, to load a single chain, or to accelerate manipulation of large models (trace and cartoon may be rendered with just the Calpha atoms available)
See documentation (1, 2) and practical examples.
Nice display of membrane proteins
The Orientation of Proteins in Membranes (University of Michigan, Lomize Group) website holds a database with the (approximate) positions of proteins embedded in membranes. The two membrane (lipid bilayer) surfaces are displayed as a set of dummy atoms added to the pdb file, red (O) for the outer side and blue (N) for the inner side.
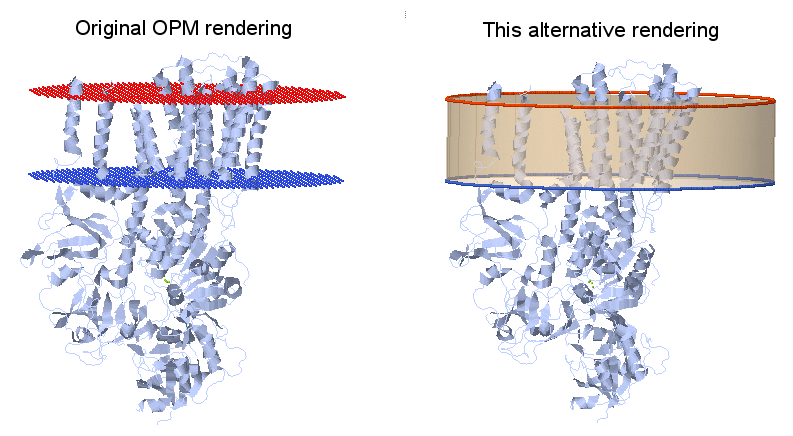
The following Jmol script locates those dummy atoms, removes them and inserts in their place the respective circular planes, making a cylinder to represent the portion of membrane where the protein is embedded. You can see the script at work at this page.
/* Code by Angel Herráez */
load "https://opm-assets.storage.googleapis.com/pdb/6xmt.pdb";
rotate X -90; /* +Z upwards, outer side */
define m~out [DUM].O; define m~in [DUM].N;
if ( {m~out}.size>0 ) {
diam~mem = 2.2 * {m~out}.x.max;
z~out = {m~out}.xyz;
if ( {m~in}.size>0 ) {
z~in = {m~in}.xyz;
} else {
z~in = - z~out;
}
} else if ( {m~in}.size>0 ) {
diam~mem = 2.2 * {m~in}.x.max;
z~in = {m~in}.xyz;
z~out = - z~in;
} else {
diam~mem = 52;
z~out = 14.9;
z~in = -14.9;
};
draw surf~mem diameter @diam~mem color translucent navajoWhite cylinder @z~in @z~out fill;
draw surf~out scale @diam~mem color orangeRed arc @z~out @z~in;
draw surf~in scale @diam~mem color royalBlue arc @z~in @z~out;
delete [DUM];
select protein; cartoon only; color chain;
Contributors
AngelHerraez, Wayne Decatur, EricMartz, NicolasVervelle, Pimpim