Difference between revisions of "Recycling Corner"
AngelHerraez (talk | contribs) (→Exporting an image from the applet) |
AngelHerraez (talk | contribs) m (→Exchange of data between JmolScript and JavaScript) |
||
| (43 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
Feel free to add your material! | Feel free to add your material! | ||
| − | |||
For example: | For example: | ||
| − | == Jmol icons == | + | == Jmol logos, banners, icons == |
| − | === New | + | === New 2013 Jmol icon and logos === |
| − | + | Icon, png: | |
| − | + | ||
| − | + | [[Image:Jmol_icon13.png]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | [[Image:Jmol_icon_13.ico.zip|Windows icon]]; .ico files cannot be shown on this webpage, but you can download this one zipped. | |
| + | for more icon formats, visit [http://biomodel.uah.es/Jmol/logos/ Angel's webpage] | ||
| − | + | Logos and banners: | |
| − | |||
| − | [[Image: | + | [[Image:Jmol_logo13.png]] |
| + | [[Image:JSmol_logo13.png]] | ||
| − | [[ | + | 'Made with Jmol' banners, in several languages: [[Jmol_Images]] |
| − | + | === Older versions === | |
| − | + | [[Jmol_Images|Old logos and icons]] | |
| − | [[Jmol_Images| | ||
| Line 70: | Line 57: | ||
For an alternative method, see [http://www.cem.msu.edu/~reusch/VirtTxtJml/strcopen.htm Bill Reusch's page] that uses innerHTML. | For an alternative method, see [http://www.cem.msu.edu/~reusch/VirtTxtJml/strcopen.htm Bill Reusch's page] that uses innerHTML. | ||
| + | For other pop-up solutions, see also [[#Opening_a_duplicate_of_the_model_in_a_resizable_pop-up_window|below]]. | ||
=== Dynamically resized panels layout === | === Dynamically resized panels layout === | ||
| Line 87: | Line 75: | ||
The code below was used to create the palettes shown above. It's a pure javascript color picker that can be used for changing color of selected part of molecule, applet background, or any feature of the model (cartoons, surfaces, etc.). | The code below was used to create the palettes shown above. It's a pure javascript color picker that can be used for changing color of selected part of molecule, applet background, or any feature of the model (cartoons, surfaces, etc.). | ||
| − | Sample of use | + | Sample of use, JavaScript code needed, example page: [[Recycling_Corner/Molecule_color_picker]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | JavaScript code needed | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== "Basic" spin toggle === | === "Basic" spin toggle === | ||
| Line 188: | Line 90: | ||
(''The word "spin" above can be changed to any other text you like.'') | (''The word "spin" above can be changed to any other text you like.'') | ||
| − | |||
=== "Foolproof" spin toggle === | === "Foolproof" spin toggle === | ||
| Line 196: | Line 97: | ||
Valid for recent versions of Jmol: | Valid for recent versions of Jmol: | ||
| − | |||
<pre> | <pre> | ||
jmolButton("if(_spinning);spin off;else;spin on;endif","toggle spin") | jmolButton("if(_spinning);spin off;else;spin on;endif","toggle spin") | ||
</pre> | </pre> | ||
| + | Or even more compact: | ||
| + | <pre> | ||
| + | jmolButton("spin @{!_spinning}","toggle spin") | ||
| + | </pre> | ||
| + | |||
(If you prefer <code>jmolLink</code>, it can be used in the same manner. If you don't use <code>Jmol.js</code>, and you have a good reason for not using it, you will know how to modify this code.) | (If you prefer <code>jmolLink</code>, it can be used in the same manner. If you don't use <code>Jmol.js</code>, and you have a good reason for not using it, you will know how to modify this code.) | ||
| Line 239: | Line 144: | ||
=== Slider controls for Jmol === | === Slider controls for Jmol === | ||
| − | + | See [[Recycling_Corner/Slider controls]] | |
| − | |||
| − | |||
| − | See | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Echoing PDB "Title" to the applet window === | === Echoing PDB "Title" to the applet window === | ||
| Line 344: | Line 186: | ||
* [http://www.marietta.edu/%7Espilatrs/MnQuiry/JmolDemo/jmolImgCopy-test.html inserted into the same web page] | * [http://www.marietta.edu/%7Espilatrs/MnQuiry/JmolDemo/jmolImgCopy-test.html inserted into the same web page] | ||
| − | + | ||
| + | === Opening a duplicate of the model in a resizable pop-up window === | ||
| + | The model, with is current state, is duplicated or cloned into a new (pop-up) window so it may be displayed at a larger size (and the user may resize that window at will). | ||
| + | |||
| + | See [[Recycling_Corner/Duplicate_Pop-up]] | ||
| + | |||
=== Style transitions === | === Style transitions === | ||
| Line 387: | Line 234: | ||
''Warning!'' | ''Warning!'' | ||
* This method has not been thoroughly tested and might fail or break the page; particularly, testing under different browsers and OS's is needed. | * This method has not been thoroughly tested and might fail or break the page; particularly, testing under different browsers and OS's is needed. | ||
| + | |||
=== Including special characters in echo === | === Including special characters in echo === | ||
| Line 393: | Line 241: | ||
echo "\u2192 is an arrow pointing right"; | echo "\u2192 is an arrow pointing right"; | ||
echo "\u00B2 is the superscript two, or square power"; | echo "\u00B2 is the superscript two, or square power"; | ||
| + | Note that, in these cases, the use of quotes surrounding the echoed text is compulsory. | ||
| + | |||
| + | Some Unicode characters may not be display in all operating systems (depending on font support). | ||
| + | For example, try these triangles and arrows: | ||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Rendered characters !! Jmol script | ||
| + | |- | ||
| + | | ◄ ► ⏴ ⏵ ◀ ▶ || <code>echo "\u25C4 \u25BA \u23F4 \u23F5 \u25C0 \u25B6"</code> | ||
| + | |- | ||
| + | | ⇤ ← → ⇥ || <code>echo "\u21E4 \u2190 \u2192 \u21E5"</code> | ||
| + | |- | ||
| + | | ⏮ ⏭ ⏪ ⏩ ⏸ ⏯ ■ ⏹ || <code>echo "\u23EE \u23ED \u23EA \u23E9 \u23F8 \u23EF \u25A0 \u23F9"</code> | ||
| + | |} | ||
| + | |||
| + | which may look like this, or different depending on OS and web browser: | ||
| + | [[File:Unicode triangle arrows.png]] | ||
=== Center of mass === | === Center of mass === | ||
Jmol centers the model around its geometric center, i.e. the center of the boundbox. If you are interested in the mass center of the molecule: | Jmol centers the model around its geometric center, i.e. the center of the boundbox. If you are interested in the mass center of the molecule: | ||
| − | + | See [[Recycling_Corner/Center_Of_Mass]] | |
| − | < | + | |
| − | function | + | |
| − | + | === RIBOZOME - an Alpha Helix Generator script === | |
| − | + | Creates an alpha helix from a user input string. | |
| − | + | ||
| + | A script for the Jmol Application and also applied to a webpage with a J(S)mol object. | ||
| + | |||
| + | See [[Recycling_Corner/Alpha_Helix_Generator]] | ||
| + | |||
| + | === POLYMERAZE - an DNA Double Helix Generator script === | ||
| + | Creates a DNA or RNA single or double helix from a user input string. | ||
| + | |||
| + | See [[Recycling_Corner/DNA_Generator]] | ||
| + | |||
| + | |||
| + | === Drawing objects === | ||
| + | ==== Cylinder ==== | ||
| + | To draw a cylinder, you need this information: | ||
| + | * two points that define the axis of the cylinder and its length | ||
| + | ** each of these may be an XYZ coordinate, an atom, the center of an atom set, or a reference to another previously drawn object | ||
| + | * the diameter of the cylinder (in Angstroms) | ||
| + | Examples (with two XYZ points): | ||
| + | // open cylinder (tube): | ||
| + | draw myCyl diameter 6.0 color translucent cyan cylinder {13 1.58 9.3} {15 1.21 15} mesh nofill; | ||
| + | // closed at both ends: | ||
| + | draw myCyl diameter 6.0 color translucent cyan cylinder {13 1.58 9.3} {15 1.21 15} fill; | ||
| + | // closed at one end: | ||
| + | draw myCyl diameter 6.0 color translucent cyan cylinder {13 1.58 9.3} {15 1.21 15} fill; | ||
| + | draw myCap diameter 6.0 color opaque cyan circle {13 1.58 9.3} {15 1.21 15}; | ||
| + | For options, see documentation for the [{{ScriptingDoc}}#draw draw] command. | ||
| + | |||
| + | ==== Ellipsoid ==== | ||
| + | Starting from these: | ||
| + | * two points that define the main axis of the ellipsoid and its length | ||
| + | ** each of these may be an XYZ coordinate, an atom, the center of an atom set | ||
| + | |||
| + | Example, with two atoms (numbers 3 and 7) at the ends of the ellipsoid: | ||
| + | atA = {atomNo=3}; | ||
| + | atB = {atomNo=7}; | ||
| + | dC = {(atA,atB)}; // defines the center, midpoint between both atoms | ||
| + | dE = 0.5; // eccentricity (<1 for cigar-shaped, 1 for a sphere, >1 for disk-shaped) | ||
| + | dR = 7; // resolution of the mesh or surface | ||
| + | dX = {atA}.x - {atB}.x; // for defining the orientation | ||
| + | dY = {atA}.y - {atB}.y; | ||
| + | dZ = {atA}.z - {atB}.z; | ||
| + | dL = {atA}.xyz.distance({atB}.xyz) /2; // length of the main ellipsoid axis | ||
| + | isosurface myEllipA color blueTint scale @dL resolution @dR center @dC ellipsoid {@dX @dY @dZ @dE} mesh nofill; | ||
| + | isosurface myEllipB color translucent 0.7 lime scale @dL resolution @dR center @dC ellipsoid {@dx @dy @dz @dE} fill; | ||
| + | Resolution and eccentricity will need to be adjusted depending on your case. | ||
| + | |||
| + | === Element legend === | ||
| + | This will work with any loaded molecule (it automatically reads the list of elements present). | ||
| + | |||
| + | ''It requires Jmol 14.6.0 or later.'' | ||
| + | |||
| + | Use either with <code>color echo black; background white; </code> | ||
| + | or with <code>color echo white; background black; </code> | ||
| + | function createElementKey() { | ||
| + | var y = 90; | ||
| + | for (var e in {*}.element.pivot) { | ||
| + | var c = {element=@e}.color; | ||
| + | draw id @{"d_"+ e} diameter 2 [90 @y %] color @c; | ||
| + | set echo id @{"e_" + e} [91 @{y-1} %]; | ||
| + | echo @e; | ||
| + | font echo 24 bold sans; | ||
| + | y -= 5; | ||
| + | } | ||
| + | } | ||
| + | createElementKey; | ||
| + | |||
| + | |||
| + | === Rainbow color key (legend) === | ||
| + | This shows (inside J(S)mol) the actual colors used by Jmol in the "roygb" scheme. | ||
| + | (there are other ways to achieve this) | ||
| + | <pre>function rainbowKey(minValue,maxValue,fontSize) { | ||
| + | rainbowColors = ['[x0000FF]','[x0020FF]','[x0040FF]','[x0060FF]','[x0080FF]', | ||
| + | '[x00A0FF]','[x00C0FF]','[x00E0FF]','[x00FFFF]','[x00FFE0]','[x00FFC0]', | ||
| + | '[x00FFA0]','[x00FF80]','[x00FF60]','[x00FF40]','[x00FF20]','[x00FF00]', | ||
| + | '[x20FF00]','[x40FF00]','[x60FF00]','[x80FF00]','[xA0FF00]','[xC0FF00]', | ||
| + | '[xE0FF00]','[xFFFF00]','[xFFE000]','[xFFC000]','[xFFA000]','[xFF8000]', | ||
| + | '[xFF6000]','[xFF4000]','[xFF2000]','[xFF0000]']; | ||
| + | background echo darkgray; | ||
| + | font echo @fontSize sans; | ||
| + | for (i=1;i<=rainbowColors.length;i++) { | ||
| + | n = "k"+i; // each echo needs a unique ID | ||
| + | y = 99 - 2*i; | ||
| + | set echo id @n [1 @y %]; | ||
| + | color echo @{rainbowColors[i]}; | ||
| + | v = (minValue + (maxValue-minValue) * i / rainbowColors.length) % 1; | ||
| + | if (v.size<4) {v = " " + v;} | ||
| + | t = "\u2B24 " + v; // circle; \u25A0 square | ||
| + | echo @t; | ||
} | } | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
} | } | ||
| + | rainbowKey(5.0,25.0,18); | ||
| + | </pre> | ||
| + | |||
| − | function | + | === Test if a file exists before displaying it === |
| − | + | ==== Using HTML, Javascript and Jmol scripting: ==== | |
| + | A Javascript function: | ||
| + | <pre>function loadFile(x) { | ||
| + | var s = 'filedat = load("' + x + '"); exists = true; '+ | ||
| + | 'if ( filedat.find("NetworkError") != 0 ) { exists = false; } '+ | ||
| + | 'elseif ( filedat.find("Not Found") != 0 ) { exists = false; } '+ | ||
| + | 'if ( exists ) { load var filedat; } else { zap; set echo e0 50% 50%; set echo e0 center; echo "This file cannot be found"; }'; | ||
| + | Jmol.script(myJmol, s); | ||
} | } | ||
| + | </pre> | ||
| + | |||
| + | And the HTML element calling it: | ||
| + | <pre><input type="button" onClick="loadFile('test.mol')" value="load it"> | ||
| + | </pre> | ||
| + | |||
| + | ==== Using Jmol callbacks to Javascript: ==== | ||
| + | Another possibility is to use [[Jmol_JavaScript_Object/Info#Callbacks|loadStructCallback]] to detect the failure in loading the file. For instance: | ||
| − | function | + | In Javascript: |
| − | + | <pre> | |
| − | + | Info.loadstructcallback = 'loadCB'; | |
| + | function loadCB() { | ||
| + | if (arguments[5]==0 && !window['noFile']) { //avoid repeated callbacks by zap | ||
| + | Jmol.script(myJmol, 'zap; set echo e0 50% 50%; set echo e0 center; echo "This file cannot be found";'); | ||
| + | window['noFile'] = true; | ||
| + | } else { | ||
| + | window['noFile'] = false; | ||
| + | } | ||
} | } | ||
</pre> | </pre> | ||
| − | + | ==== Using only Jmol scripting: ==== | |
| − | + | Javascript: | |
| − | <pre> | + | <pre>Info.script = 'function loadFile(x) { filedat=load(x); exists=true; '+ |
| − | + | 'if (filedat.find("NetworkError")!=0) {exists=false;} elseif (filedat.find("Not Found")!=0) {exists=false;}; '+ | |
| − | + | 'if (exists) {load var filedat;} else {zap; set echo e0 50% 50%; set echo e0 center; echo "This file cannot be found";} }; '; | |
| − | + | </pre> | |
| + | |||
| + | And HTML: | ||
| + | <pre> | ||
| + | <p>This file exists in the same folder | ||
| + | <script type="text/javascript">Jmol.jmolButton(myJmol, "loadFile('test.mol')", "load it");</script></p> | ||
| + | <p>This file does not exist | ||
| + | <script type="text/javascript">Jmol.jmolButton(myJmol, "loadFile('test2.mol')", "load it");</script></p> | ||
| + | </pre> | ||
| + | |||
| + | ==== Using an AJAX call: ==== | ||
| + | Javascript: | ||
| + | <pre>function loadFile(pathFilenameExt) { | ||
| + | var req = new XMLHttpRequest(); | ||
| + | req.onreadystatechange = function() { | ||
| + | if (this.readyState != 4) { return; } /*not finished yet*/ | ||
| + | if (this.status == 200) { | ||
| + | Jmol.script(myJmol, 'load "' + pathFilenameExt + '";'); | ||
| + | } else { | ||
| + | Jmol.script(myJmol, 'zap; set echo e0 50% 50%; set echo e0 center; echo "This file cannot be found";'); | ||
| + | } | ||
| + | }; | ||
| + | req.open('GET', pathFilenameExt, false); | ||
| + | req.send(); | ||
| + | } | ||
</pre> | </pre> | ||
| − | + | ||
| − | <pre> | + | And the HTML: |
| − | + | <pre><p>This file exists in the same folder <input type="button" onClick="loadFile('test.mol')" value="load it"></p> | |
| + | <p>This file does not exist <input type="button" onClick="loadFile('test2.mol')" value="load it"></p> | ||
</pre> | </pre> | ||
| − | + | ||
| − | <pre> | + | ''(Based on a contribution by Bilal Nizami)'' |
| − | + | ||
| + | === Improved lighting === | ||
| + | The lighting of atoms, bonds and ribbons and the thickness of ribbons may be customized to provide a nicer look, by adjusting several settings. Here is an example: | ||
| + | |||
| + | [[File:Fast, fancy, shiny.png]] | ||
| + | |||
| + | [[File:Fast, fancy, shiny ball-and-stick.png]] | ||
| + | |||
| + | Default: | ||
| + | reset lighting; set cartoonsFancy off; set hermiteLevel 0; set antialiasDisplay off; | ||
| + | |||
| + | "Fast": (quick rendering, same as default except antialias is applied for a better comparison with the other two) | ||
| + | reset lighting; set cartoonsFancy off; set hermiteLevel 0; set antialiasDisplay on; | ||
| + | |||
| + | "Fancy": (relevant only for cartoons, without effect on atoms and bonds) | ||
| + | reset lighting; set cartoonsFancy on; set hermiteLevel 20; set antialiasDisplay on; | ||
| + | |||
| + | "Shiny": (adds light and shadow effects) | ||
| + | (more similar to rendering in other software like e.g. Chimera) | ||
| + | set ambientPercent 25; set specularPower 100; set specularPercent 22; set specularExponent 4; set cartoonsFancy on; set antialiasDisplay on; set hermiteLevel 20; | ||
| + | |||
| + | [[File:Shiny-rendering-spacefill.png]] | ||
| + | |||
| + | Note: Jmol defaults may be restored using <code>reset lighting</code> which applies these values: | ||
| + | {| style="border-spacing:8px 0;" | ||
| + | |- | ||
| + | || set | ||
| + | || ambientPercent | ||
| + | || diffusePercent | ||
| + | || specular | ||
| + | || specularPercent | ||
| + | || specularPower | ||
| + | || specularExponent | ||
| + | |- | ||
| + | | align=center| default value: | ||
| + | | align=center| 45 | ||
| + | | align=center| 84 | ||
| + | | align=center| on | ||
| + | | align=center| 22 | ||
| + | | align=center| 40 | ||
| + | | align=center| 6 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | {| style="border-spacing:8px 0; margin-top:1.4em;" | ||
| + | |- | ||
| + | || set | ||
| + | || zDepth | ||
| + | || zSlab | ||
| + | || zShade | ||
| + | || zShadePower | ||
| + | || celShading | ||
| + | || celShadingPower | ||
| + | |- style="" | ||
| + | | align=center| default value: | ||
| + | | align=center| 0 | ||
| + | | align=center| 50 | ||
| + | | align=center| off | ||
| + | | align=center| 3 | ||
| + | | align=center| off | ||
| + | | align=center| 10 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | For more details about each of these settings, see the | ||
| + | [{{ScriptingDoc}}#setlighting Scripting Documentation]. | ||
| + | |||
| + | === Extract phi and psi values === | ||
| + | The dihedral angles phi and psi are known in Jmol, as properties of each amino acid residue in a protein, which are copied to properties of each atom in that residue. | ||
| + | |||
| + | These scripts may be used to extract their values into an array: | ||
| + | |||
| + | <table> | ||
| + | <tr> | ||
| + | <td style="vertical-align:top;padding-right:2em;">For the first polymer in the first model in the loaded file: | ||
| + | p = getProperty("polymerinfo.models[1].polymers[1].monomers").select("psi,phi") | ||
| + | p will be an associative array, one element per residue, each one with keys "phi" and "psi" and their values. | ||
| + | </td> | ||
| + | <td>Result for the first 2 and last 2 residues: | ||
| + | print p | ||
| + | { | ||
| + | "psi" : 135.30487 | ||
| + | } | ||
| + | { | ||
| + | "phi" : -103.61969 | ||
| + | "psi" : 110.173744 | ||
| + | } | ||
| + | ... | ||
| + | { | ||
| + | "phi" : -81.423874 | ||
| + | "psi" : 131.89566 | ||
| + | } | ||
| + | { | ||
| + | "phi" : -166.42226 | ||
| + | } | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | <table> | ||
| + | <tr> | ||
| + | <td style="vertical-align:top;padding-right:2em;">Another way, which will read all protein chains present: | ||
| + | <pre>ph = []; | ||
| + | ps = []; | ||
| + | n1 = {protein}.resno.min; | ||
| + | n2 = {protein}.resno.max; | ||
| + | for (r=n1; r<=n2; r++) { | ||
| + | ph.push({resno=r}.phi); | ||
| + | ps.push({resno=r}.psi); | ||
| + | } | ||
</pre> | </pre> | ||
| + | </td> | ||
| + | <td>Result: | ||
| + | print ph.join("\n") | ||
| + | NaN | ||
| + | -34.539898 | ||
| + | ... | ||
| + | -81.423874 | ||
| + | -166.42226 | ||
| + | |||
| + | print ps.join("\n") | ||
| + | 135.30487 | ||
| + | 36.724583 | ||
| + | ... | ||
| + | 131.89566 | ||
| + | NaN | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | === Front of the molecule, selecting (by percentage) === | ||
| + | |||
| + | The Jmol function at [[Recycling_Corner/Front_Selection]] selects a specified percentage of the front of the molecule, according to the orientation of the molecule when the function is executed. | ||
| + | |||
| + | === Exchange of data between JmolScript and JavaScript === | ||
| − | < | + | # Passing a variable from JavaScript to a JSmol variable: |
| + | #* If the JS variable is e.g. myJSvar = 55, doing this will assign its value to a variable in JSmol: | ||
| + | #: <code>myJSmolVar = javascript("myJSvar"); print myJSmolVar; </code> // prints 55 | ||
| + | # Passing a variable from JSmol to a JavaScript variable: | ||
| + | #: <code>myJSvar = Jmol.evaluateVar(myJSmol, myJSmolVar) </code>. Instead of the name of the variable, in the position of myJSmolVar you may use a Jmol math expression or some information from the molecule. <code>myJSmol</code> is the id/name of the JSmol object in the webpage (e.g. myJmol or jmolApplet0). [[Jmol_JavaScript_Object/Functions#evaluateVar|More details about evaluateVar()]]. | ||
== Custom pop-up menus == | == Custom pop-up menus == | ||
| − | + | Jmol (both application and applet) allows a customized pop-up menu instead of the standard default menu. | |
Their definition and use are explained in [[Custom Menus]]. | Their definition and use are explained in [[Custom Menus]]. | ||
Examples: | Examples: | ||
| − | * Default menu is built using this: [ | + | * Default menu is built using this: [{{SVN Trunk}}Jmol/jmol.mnu jmol.mnu] |
* An example of custom menu: [[test.mnu]] | * An example of custom menu: [[test.mnu]] | ||
Latest revision as of 18:23, 2 September 2024
This section intends to collect material by some users that may be useful to other users.
Feel free to add your material!
For example:
Contents
- 1 Jmol logos, banners, icons
- 2 Templates for creating Jmol webpages
- 3 Reusable scripts
- 3.1 Molecule color picker
- 3.2 "Basic" spin toggle
- 3.3 "Foolproof" spin toggle
- 3.4 Slider controls for Jmol
- 3.5 Echoing PDB "Title" to the applet window
- 3.6 Providing a 'please wait' notice while the applet loads
- 3.7 Exporting an image from the applet
- 3.8 Opening a duplicate of the model in a resizable pop-up window
- 3.9 Style transitions
- 3.10 Replacing applet with signed applet upon demand
- 3.11 Including special characters in echo
- 3.12 Center of mass
- 3.13 RIBOZOME - an Alpha Helix Generator script
- 3.14 POLYMERAZE - an DNA Double Helix Generator script
- 3.15 Drawing objects
- 3.16 Element legend
- 3.17 Rainbow color key (legend)
- 3.18 Test if a file exists before displaying it
- 3.19 Improved lighting
- 3.20 Extract phi and psi values
- 3.21 Front of the molecule, selecting (by percentage)
- 3.22 Exchange of data between JmolScript and JavaScript
- 4 Custom pop-up menus
Jmol logos, banners, icons
New 2013 Jmol icon and logos
Icon, png:
File:Jmol icon 13.ico.zip; .ico files cannot be shown on this webpage, but you can download this one zipped.
for more icon formats, visit Angel's webpage
Logos and banners:
'Made with Jmol' banners, in several languages: Jmol_Images
Older versions
Templates for creating Jmol webpages
Jmol_Web_Page_Maker
Jonathan Gutow has written a Java program that automates generation of webpages that include Jmol. The two templates below are among the types of web pages that can be generated. When using Jmol_Web_Page_Maker for the templates below, you can choose the view(s) to be displayed inside the Jmol application and have the necessary scripts, images plus .html code generated and saved automatically. Visit his website.
This is now superceeded by the "export to web page" ability included within recent versions of Jmol application (11.3.x).
Easy assignments for students or quick building of a simple page!
SChiSM2
SChiSM2 is a program running in a web server that lets you easily create a web page that includes interactive molecular models using JmolApplet.
- SChiSM2 home page
- A description has been published:
- S. Cammer (2007) SChiSM2: creating interactive web page annotations of molecular structure models using Jmol. Bioinformatics 23:383-384. doi:10.1093/bioinformatics/btl603 [1]
Jmol "pop-up" and "pop-in"
Delays the appearance of Jmol panel until the user requests it. An image or caption is shown and upon user's choice it is replaced by the applet.
No more waiting for Java and Jmol to load if you are not interested in the 3D model!
Two methods have been developed:
- Method A - versatile: The user clicks to see the 3D model, and chooses whether it will be opened inline within the page (âpop-inâ), or in a common pop-up window, or in separate pop-up windows for each model. (Uses <iframe>.)
- Method B - simpler: The user clicks to see the 3D model, always inline within the page (âpop-inâ). (Uses <div>.) (A similar method is used on Jmol home webpage.)
You can find more details, demo pages and downloadable templates for both methods at Biomodel website.
For an alternative method, see Bill Reusch's page that uses innerHTML.
For other pop-up solutions, see also below.
Dynamically resized panels layout
Six templates for dividing the screen into two or more panels, either for Jmol or for content, all sized in percent relative to window size, using the whole window space, and automatically resized when the window is resized; no scrollbars in Jmol section, scrollbars in content if needed, no duplicate parallel scrollbars.
No more guessing the user's screen resolution, browser chrome, default font size!
You can find more details, demo pages and downloadable templates at Biomodel website.
Reusable scripts
Molecule color picker
The code below was used to create the palettes shown above. It's a pure javascript color picker that can be used for changing color of selected part of molecule, applet background, or any feature of the model (cartoons, surfaces, etc.).
Sample of use, JavaScript code needed, example page: Recycling_Corner/Molecule_color_picker
"Basic" spin toggle
Quite trivial, but compact and useful. However, if other scripts, controls, or the user change the spin status, the checkbox will need to be reset accordingly using javascript. (To avoid this limitation, see the "foolproof" solution below)
It will work with any version of Jmol (as long as Jmol.js is being used in the web page).
jmolCheckbox("spin on", "spin off", "spin", false)
if the model is initially not spinning (the default);
or
jmolCheckbox("spin on", "spin off", "spin", true)
if the model is initially spinning (due to another script).
(The word "spin" above can be changed to any other text you like.)
"Foolproof" spin toggle
Reads the current spin status from the applet and changes it, so it can cope with any other previous change via script, popup menu, page controls, etc.
The best and simplest trick:
Valid for recent versions of Jmol:
jmolButton("if(_spinning);spin off;else;spin on;endif","toggle spin")
Or even more compact:
jmolButton("spin @{!_spinning}","toggle spin")
(If you prefer jmolLink, it can be used in the same manner. If you don't use Jmol.js, and you have a good reason for not using it, you will know how to modify this code.)
(The words "toggle spin" above can be changed to any other text you like.)
The old trick:
Only compatible with Jmol 11.x (as long as Jmol.js is being used in the web page).
jmolButton("show spin", "toggle spin")
(If you prefer jmolLink, it can be used in the same manner.)
(The words "toggle spin" above can be changed to any other text you like.)
You must include these additional pieces of code:
Somewhere between <script> tags (recommended: within the head section):
function messageProcess(a_,b_)
{ // Convert both parameters from Java strings to JavaScript strings:
var a = (""+a_).substring("jmolApplet".length)
var b = ""+b_
// a_ will start with "jmolApplet": strip and leave just the ID part
/* SPIN DETECTION AND TOGGLE */
if ( b.indexOf("set spin") != -1 )
{ if ( b.indexOf("spin on") != -1 ) //was spinning
{ jmolScript("spin off", a)
}
else //was not spinning
{ jmolScript("spin on", a)
}
}
/* END of SPIN DETECTION AND TOGGLE */
}
and somewhere between <script> tags, after the call to jmolInitialize() and before the call to jmolApplet():
jmolSetCallback("messageCallback", "messageProcess")
(The word "messageProcess" above can be changed to any other text you like, as long as it is the same in the above two sections of code.)
Slider controls for Jmol
See Recycling_Corner/Slider controls
Echoing PDB "Title" to the applet window
The following JavaScript code snippet will report the title of a PDB entry to the applet window when a PDB file is loaded. This should be included within a JavaScript portion of the html file that loads the applet and the PDB file. One caveat: the "TITLE" in a PDB file is depositor-generated and may or may not be "useful" to the end user! :)
var headerInfo = jmolGetPropertyAsString("fileHeader");
var cutUp = headerInfo.split("\n");
var headerstring="";
for (l=0;l<cutUp.length;l++) {
var regexp = new RegExp("TITLE.{5}(.*)\s*");
var temp = cutUp[l];
if (temp.search(regexp) == 0) {
temp2 = RegExp.$1;
temp2.replace(/^\s+|\s+$/g, "").replace(/\s+/g, " ");
temp2 = temp2 + "|";
headerstring += temp2; }
}
}
headerstring.replace(/TITLE/, "");
jmolScript("set echo depth 0; set echo headerecho 2% 2%; font echo 12 sanserif bolditalic; color echo green");
jmolScript('echo "' + headerstring + '"');
Bob Hanson has also contributed this much more elegant way to accomplish the same thing all within JmolScript:
function getHeader()
var titleLines = getProperty("fileHeader").split().find("TITLE").split()
for (var i = 1; i <= titleLines.size;i = i + 1)
titleLines[i] = (titleLines[i])[11][0].trim()
end for
return titleLines.join("|")
end function
set echo bottom left
echo @{getheader()}
Providing a 'please wait' notice while the applet loads
in a separate page
Exporting an image from the applet
An image of the current view may be exported from the Jmol applet, either
- in a pop-up window, also compatible with JSO and JSmol.
- inserted into the same web page
Opening a duplicate of the model in a resizable pop-up window
The model, with is current state, is duplicated or cloned into a new (pop-up) window so it may be displayed at a larger size (and the user may resize that window at will).
See Recycling_Corner/Duplicate_Pop-up
Style transitions
Smooth transitions from one rendering style to another.
Spacefill to ball & stick
for (var i=0;i<=9;i++) {
s=100-80*i/9;
spacefill @s%;
w=0.9-0.75*i/9;
wireframe @w;
delay 0.2;
}
Can be given in a single line:
for(var i=0;i<=9;i++){s=100-80*i/9;spacefill @s%;w=0.9-0.75*i/9;wireframe @w;delay 0.2;}
Replacing applet with signed applet upon demand
The signed applet has certain capabilities that the unsigned applet lacks, but using the signed applet will raise some dialogs with security warnings that the general user may find scary and, if permission is denied, the expanded features will not be available and hence the page will fail to do what is expected.
This may lead to a situation where webpage authors wish to use the unsigned applet by default and make the switch to the signed applet if and when the users requests a certain capability (like saving a model to disk). The code here will do that.
The rationale is:
- Store the current state of the model in the applet so that it can be restored after the applet switch.
- Destroy the applet object.
- Insert the signed applet in the same place.
- Load back the model and state.
Latest Jmol.js (post-26 Sep 2011, Jmol_12.2.RC8 or later) includes a new jmolSwitchToSignedApplet() function to do this replacement easier:
The code (html + JavaScript, relying on Jmol.js):
<script type="text/javascript">
jmolInitialize(".");
jmolApplet(400, 'load example.mol;', 'ABC');
</script>
<br>
<input type="button" value="make the switch" onClick="jmolSwitchToSignedApplet('ABC')">
That's all! Note that 'ABC' is an example of an applet suffix ID, and if omitted will default to '0' (zero) in both jmolApplet() and jmolSwitchToSignedApplet().
Warning!
- This method has not been thoroughly tested and might fail or break the page; particularly, testing under different browsers and OS's is needed.
Including special characters in echo
Non-keyboard characters may be included in echo texts using their Unicode character number, like this:
echo "\u03B1 is the alpha letter"; echo "\u2192 is an arrow pointing right"; echo "\u00B2 is the superscript two, or square power";
Note that, in these cases, the use of quotes surrounding the echoed text is compulsory.
Some Unicode characters may not be display in all operating systems (depending on font support). For example, try these triangles and arrows:
| Rendered characters | Jmol script |
|---|---|
| ◄ ► ⏴ ⏵ ◀ ▶ | echo "\u25C4 \u25BA \u23F4 \u23F5 \u25C0 \u25B6"
|
| ⇤ ← → ⇥ | echo "\u21E4 \u2190 \u2192 \u21E5"
|
| ⏮ ⏭ ⏪ ⏩ ⏸ ⏯ ■ ⏹ | echo "\u23EE \u23ED \u23EA \u23E9 \u23F8 \u23EF \u25A0 \u23F9"
|
which may look like this, or different depending on OS and web browser:
Center of mass
Jmol centers the model around its geometric center, i.e. the center of the boundbox. If you are interested in the mass center of the molecule:
See Recycling_Corner/Center_Of_Mass
RIBOZOME - an Alpha Helix Generator script
Creates an alpha helix from a user input string.
A script for the Jmol Application and also applied to a webpage with a J(S)mol object.
See Recycling_Corner/Alpha_Helix_Generator
POLYMERAZE - an DNA Double Helix Generator script
Creates a DNA or RNA single or double helix from a user input string.
See Recycling_Corner/DNA_Generator
Drawing objects
Cylinder
To draw a cylinder, you need this information:
- two points that define the axis of the cylinder and its length
- each of these may be an XYZ coordinate, an atom, the center of an atom set, or a reference to another previously drawn object
- the diameter of the cylinder (in Angstroms)
Examples (with two XYZ points):
// open cylinder (tube):
draw myCyl diameter 6.0 color translucent cyan cylinder {13 1.58 9.3} {15 1.21 15} mesh nofill;
// closed at both ends:
draw myCyl diameter 6.0 color translucent cyan cylinder {13 1.58 9.3} {15 1.21 15} fill;
// closed at one end:
draw myCyl diameter 6.0 color translucent cyan cylinder {13 1.58 9.3} {15 1.21 15} fill;
draw myCap diameter 6.0 color opaque cyan circle {13 1.58 9.3} {15 1.21 15};
For options, see documentation for the draw command.
Ellipsoid
Starting from these:
- two points that define the main axis of the ellipsoid and its length
- each of these may be an XYZ coordinate, an atom, the center of an atom set
Example, with two atoms (numbers 3 and 7) at the ends of the ellipsoid:
atA = {atomNo=3};
atB = {atomNo=7};
dC = {(atA,atB)}; // defines the center, midpoint between both atoms
dE = 0.5; // eccentricity (<1 for cigar-shaped, 1 for a sphere, >1 for disk-shaped)
dR = 7; // resolution of the mesh or surface
dX = {atA}.x - {atB}.x; // for defining the orientation
dY = {atA}.y - {atB}.y;
dZ = {atA}.z - {atB}.z;
dL = {atA}.xyz.distance({atB}.xyz) /2; // length of the main ellipsoid axis
isosurface myEllipA color blueTint scale @dL resolution @dR center @dC ellipsoid {@dX @dY @dZ @dE} mesh nofill;
isosurface myEllipB color translucent 0.7 lime scale @dL resolution @dR center @dC ellipsoid {@dx @dy @dz @dE} fill;
Resolution and eccentricity will need to be adjusted depending on your case.
Element legend
This will work with any loaded molecule (it automatically reads the list of elements present).
It requires Jmol 14.6.0 or later.
Use either with color echo black; background white;
or with color echo white; background black;
function createElementKey() {
var y = 90;
for (var e in {*}.element.pivot) {
var c = {element=@e}.color;
draw id @{"d_"+ e} diameter 2 [90 @y %] color @c;
set echo id @{"e_" + e} [91 @{y-1} %];
echo @e;
font echo 24 bold sans;
y -= 5;
}
}
createElementKey;
Rainbow color key (legend)
This shows (inside J(S)mol) the actual colors used by Jmol in the "roygb" scheme. (there are other ways to achieve this)
function rainbowKey(minValue,maxValue,fontSize) {
rainbowColors = ['[x0000FF]','[x0020FF]','[x0040FF]','[x0060FF]','[x0080FF]',
'[x00A0FF]','[x00C0FF]','[x00E0FF]','[x00FFFF]','[x00FFE0]','[x00FFC0]',
'[x00FFA0]','[x00FF80]','[x00FF60]','[x00FF40]','[x00FF20]','[x00FF00]',
'[x20FF00]','[x40FF00]','[x60FF00]','[x80FF00]','[xA0FF00]','[xC0FF00]',
'[xE0FF00]','[xFFFF00]','[xFFE000]','[xFFC000]','[xFFA000]','[xFF8000]',
'[xFF6000]','[xFF4000]','[xFF2000]','[xFF0000]'];
background echo darkgray;
font echo @fontSize sans;
for (i=1;i<=rainbowColors.length;i++) {
n = "k"+i; // each echo needs a unique ID
y = 99 - 2*i;
set echo id @n [1 @y %];
color echo @{rainbowColors[i]};
v = (minValue + (maxValue-minValue) * i / rainbowColors.length) % 1;
if (v.size<4) {v = " " + v;}
t = "\u2B24 " + v; // circle; \u25A0 square
echo @t;
}
}
rainbowKey(5.0,25.0,18);
Test if a file exists before displaying it
Using HTML, Javascript and Jmol scripting:
A Javascript function:
function loadFile(x) {
var s = 'filedat = load("' + x + '"); exists = true; '+
'if ( filedat.find("NetworkError") != 0 ) { exists = false; } '+
'elseif ( filedat.find("Not Found") != 0 ) { exists = false; } '+
'if ( exists ) { load var filedat; } else { zap; set echo e0 50% 50%; set echo e0 center; echo "This file cannot be found"; }';
Jmol.script(myJmol, s);
}
And the HTML element calling it:
<input type="button" onClick="loadFile('test.mol')" value="load it">
Using Jmol callbacks to Javascript:
Another possibility is to use loadStructCallback to detect the failure in loading the file. For instance:
In Javascript:
Info.loadstructcallback = 'loadCB';
function loadCB() {
if (arguments[5]==0 && !window['noFile']) { //avoid repeated callbacks by zap
Jmol.script(myJmol, 'zap; set echo e0 50% 50%; set echo e0 center; echo "This file cannot be found";');
window['noFile'] = true;
} else {
window['noFile'] = false;
}
}
Using only Jmol scripting:
Javascript:
Info.script = 'function loadFile(x) { filedat=load(x); exists=true; '+
'if (filedat.find("NetworkError")!=0) {exists=false;} elseif (filedat.find("Not Found")!=0) {exists=false;}; '+
'if (exists) {load var filedat;} else {zap; set echo e0 50% 50%; set echo e0 center; echo "This file cannot be found";} }; ';
And HTML:
<p>This file exists in the same folder
<script type="text/javascript">Jmol.jmolButton(myJmol, "loadFile('test.mol')", "load it");</script></p>
<p>This file does not exist
<script type="text/javascript">Jmol.jmolButton(myJmol, "loadFile('test2.mol')", "load it");</script></p>
Using an AJAX call:
Javascript:
function loadFile(pathFilenameExt) {
var req = new XMLHttpRequest();
req.onreadystatechange = function() {
if (this.readyState != 4) { return; } /*not finished yet*/
if (this.status == 200) {
Jmol.script(myJmol, 'load "' + pathFilenameExt + '";');
} else {
Jmol.script(myJmol, 'zap; set echo e0 50% 50%; set echo e0 center; echo "This file cannot be found";');
}
};
req.open('GET', pathFilenameExt, false);
req.send();
}
And the HTML:
<p>This file exists in the same folder <input type="button" onClick="loadFile('test.mol')" value="load it"></p>
<p>This file does not exist <input type="button" onClick="loadFile('test2.mol')" value="load it"></p>
(Based on a contribution by Bilal Nizami)
Improved lighting
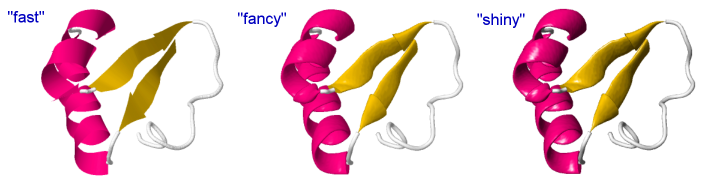
The lighting of atoms, bonds and ribbons and the thickness of ribbons may be customized to provide a nicer look, by adjusting several settings. Here is an example:
Default:
reset lighting; set cartoonsFancy off; set hermiteLevel 0; set antialiasDisplay off;
"Fast": (quick rendering, same as default except antialias is applied for a better comparison with the other two)
reset lighting; set cartoonsFancy off; set hermiteLevel 0; set antialiasDisplay on;
"Fancy": (relevant only for cartoons, without effect on atoms and bonds)
reset lighting; set cartoonsFancy on; set hermiteLevel 20; set antialiasDisplay on;
"Shiny": (adds light and shadow effects) (more similar to rendering in other software like e.g. Chimera)
set ambientPercent 25; set specularPower 100; set specularPercent 22; set specularExponent 4; set cartoonsFancy on; set antialiasDisplay on; set hermiteLevel 20;
Note: Jmol defaults may be restored using reset lighting which applies these values:
| set | ambientPercent | diffusePercent | specular | specularPercent | specularPower | specularExponent |
| default value: | 45 | 84 | on | 22 | 40 | 6 |
| set | zDepth | zSlab | zShade | zShadePower | celShading | celShadingPower |
| default value: | 0 | 50 | off | 3 | off | 10 |
For more details about each of these settings, see the Scripting Documentation.
Extract phi and psi values
The dihedral angles phi and psi are known in Jmol, as properties of each amino acid residue in a protein, which are copied to properties of each atom in that residue.
These scripts may be used to extract their values into an array:
For the first polymer in the first model in the loaded file:
p = getProperty("polymerinfo.models[1].polymers[1].monomers").select("psi,phi")
p will be an associative array, one element per residue, each one with keys "phi" and "psi" and their values. |
Result for the first 2 and last 2 residues:
print p
{
"psi" : 135.30487
}
{
"phi" : -103.61969
"psi" : 110.173744
}
...
{
"phi" : -81.423874
"psi" : 131.89566
}
{
"phi" : -166.42226
}
|
Another way, which will read all protein chains present:
ph = [];
ps = [];
n1 = {protein}.resno.min;
n2 = {protein}.resno.max;
for (r=n1; r<=n2; r++) {
ph.push({resno=r}.phi);
ps.push({resno=r}.psi);
}
|
Result:
print ph.join("\n")
NaN
-34.539898
...
-81.423874
-166.42226
print ps.join("\n")
135.30487
36.724583
...
131.89566
NaN
|
Front of the molecule, selecting (by percentage)
The Jmol function at Recycling_Corner/Front_Selection selects a specified percentage of the front of the molecule, according to the orientation of the molecule when the function is executed.
Exchange of data between JmolScript and JavaScript
- Passing a variable from JavaScript to a JSmol variable:
- If the JS variable is e.g. myJSvar = 55, doing this will assign its value to a variable in JSmol:
myJSmolVar = javascript("myJSvar"); print myJSmolVar;// prints 55
- Passing a variable from JSmol to a JavaScript variable:
myJSvar = Jmol.evaluateVar(myJSmol, myJSmolVar). Instead of the name of the variable, in the position of myJSmolVar you may use a Jmol math expression or some information from the molecule.myJSmolis the id/name of the JSmol object in the webpage (e.g. myJmol or jmolApplet0). More details about evaluateVar().
Jmol (both application and applet) allows a customized pop-up menu instead of the standard default menu. Their definition and use are explained in Custom Menus.
Examples:
Users, if you design a new menu, please, share it here.
SimpleBio
This is devised to ease the handling of the menu by inexperienced users with only basic tasks, in tutorial pages of simple biomolecules.
It includes just these entries:
- Style - with simplified contents.
- Color - compacted to have less submenu nesting.
- Spin - just with on/off, no axes or speed options.
- Select - a bit simplified.
- Advanced (*) - a new entry including:
- Choose menu (*) - that allows to revert to the standard full menu.
- Language
- Console
- About
(*) These are new entries, and so will not be localized; you may wish to edit them for your intended language.
You can get this custom menu here.
SimpleChem
This is devised to ease the handling of the menu by inexperienced users with only basic tasks, in tutorial pages of organic/inorganic molecules (no crystallography, symmetry, vibration or animation, no pdb-related options).
It includes just these entries:
- Style - with simplified contents.
- Color - simplified.
- Spin - just with on/off, no axes or speed options.
- Select - a bit simplified.
- Measurements
- Advanced (*) - a new entry including:
- Choose menu (*) - that allows to revert to the standard full menu.
- Language
- Console
- About
(*) These are new entries, and so will not be localized; you may wish to edit them for your intended language.
You can get this custom menu here.
Contributors
AngelHerraez, Ceroni, Remig, Tstout, EricMartz, NicolasVervelle, Pimpim, Gutow