Difference between revisions of "Jmol Applet"
(→Setting up a Jmol site in The Cloud, in an AWS S3 bucket) |
AngelHerraez (talk | contribs) (tips to location of applet files and Jmol.js in new distributions) |
||
| Line 34: | Line 34: | ||
==== The Jmol.js library ==== | ==== The Jmol.js library ==== | ||
For Jmol 12 (and Jmol 13, if desired), the Jmol.js library provides a set of simple scripting commands that allow rapid construction of web pages that include user interactivity such as buttons, checkboxes, links, and menus. Note that all functions in Jmol.js have counterparts in '''Jmol-JSO'''. For an overview of these functions, see the [{{Website}}jslibrary/ Jmol.js online documentation] or, for the most up-to-date info, just browse the Jmol.js source file. | For Jmol 12 (and Jmol 13, if desired), the Jmol.js library provides a set of simple scripting commands that allow rapid construction of web pages that include user interactivity such as buttons, checkboxes, links, and menus. Note that all functions in Jmol.js have counterparts in '''Jmol-JSO'''. For an overview of these functions, see the [{{Website}}jslibrary/ Jmol.js online documentation] or, for the most up-to-date info, just browse the Jmol.js source file. | ||
| + | |||
| + | ''Note:'' {{file|Jmol.js}} is phased out in Jmol v.13, in favour of the new set of object-oriented JavaScript method and {{file|.js}} files (described next). | ||
| + | Also, the download for Jmol version 13.2 or later does not include this file. If you still want to use it, you can either copy it from a former Jmol version or get it from the [{{SVN Trunk}}Jmol/appletweb/old/Jmol.js Jmol repository]. | ||
=== Jmol Applet JAR files === | === Jmol Applet JAR files === | ||
Revision as of 13:32, 25 October 2013
- Jmol as a standalone application.
- J(S)mol as a web applet or HTML5 object.
- compatibility with JavaScript frameworks or libraries.
- J(S)mol embedded in wikis or blogs.
- Borrowed JSmol, or running JSmol without your own server.
- Running Jmol using Java Web Start.
- JSmol in a Jupyter notebook.
- Jmol in an Android tablet.
- Troubleshooting.
Contents
- 1 Jmol object
- 2 Installing Jmol objects
- 3 Jmol in Wikis
- 4 Unsigned vs. Signed Applet & Data Access
- 5 Tools for Authoring Jmol Web Pages
- 6 Saving Files from the Applet
- 7 Setting up a Jmol site in The Cloud, in an AWS S3 bucket
Jmol object
Jmol Applet
Java applet version of Jmol which allows Jmol to be embedded in web pages (given that viewers have a Java plugin installed). Like with the application, commands can be applied to the model using the Scripting language. For some demonstration pages see http://jmol.sourceforge.net/demo/. To see the HTML source use your web browser View → Page Source.
Jmol HTML5 Object (JSmol)
Starting with version 13.1, Jmol may also be inserted into web pages using the non-Java alternative, called JSmol. This allows to use an HTML5 object that only requires JavaScript and has the same set of features as Jmol applet. See Jmol JavaScript Object for more details.
Installing Jmol objects
If you only wish to create a web page(s) containing molecular scenes in Jmol, there are easier ways than starting from scratch: see #Tools for Authoring Jmol Web Pages. If you wish to learn how to create web pages containing Jmol from scratch, read on.
- Get the files from SourceForge.
- Support and operating systems.
- Users: how to install Jmol application for local, standalone use.
- Web page authors: how to install Jmol objects (Jmol / JSmol) embedded within your web pages.
- Installing and Enabling Java (Proteopedia wiki). Has step by step instructions for both Windows and Mac OS X, and for the 5 most popular web browsers.
- Increasing java memory for the applet.
- Web site administrators:
- How to install the application on a web server to be delivered using Java Web Start.
- How to configure a wiki so that it uses the Jmol applet.
- Developers: compile the latest Jmol source code via subversion access.
Download the Jmol package (either .zip or tar.gz format) and extract/uncompress only the necessary files to the folder of your choice.
Which are those necessary files? Read subsections below.
To use the Jmol applet within local web pages, e.g. on the hard disk or on CD, see Jmol Applet Deployment Local for some precautions needed.
For some specific needs, you must use other files; read the sections below about the signed applet.
If the applet does not load, see also Solving Java Problems.
If you distribute your web page files, you should also include the copyright, license and information files: ![]() COPYRIGHT.txt,
COPYRIGHT.txt, ![]() LICENSE.txt and
LICENSE.txt and ![]() README.txt
README.txt
Inserting Jmol objects into web pages
Inclusion of Jmol applets/objects and interaction with them are managed easily using one of two JavaScript-based solutions:
Jmol JavaScript Object (Jmol-JSO)
Jmol-JSO (new in Jmol v.13) provides a clean, efficient way to interact with the Jmol applet from JavaScript and, in addition, allows extension to Java- and applet-challenged platforms such as the iPad, the iPhone, and Android mobile devices, and adds easy access to public databases such as the RCSB PDB database, the National Cancer Institute CACTVS server, and PubChem, even when the Jmol Java applet is not available because of a user's platform. Basic high-level scripting for these non-Java alternatives is included. In addition, Jmol-JSO adds synchronization with the JSpecView applet and 2D-to-3D model building and interconversion using the JME applet. See more details on Jmol JavaScript Object.
The Jmol.js library
For Jmol 12 (and Jmol 13, if desired), the Jmol.js library provides a set of simple scripting commands that allow rapid construction of web pages that include user interactivity such as buttons, checkboxes, links, and menus. Note that all functions in Jmol.js have counterparts in Jmol-JSO. For an overview of these functions, see the Jmol.js online documentation or, for the most up-to-date info, just browse the Jmol.js source file.
Note: ![]() Jmol.js is phased out in Jmol v.13, in favour of the new set of object-oriented JavaScript method and
Jmol.js is phased out in Jmol v.13, in favour of the new set of object-oriented JavaScript method and ![]() .js files (described next).
Also, the download for Jmol version 13.2 or later does not include this file. If you still want to use it, you can either copy it from a former Jmol version or get it from the Jmol repository.
.js files (described next).
Also, the download for Jmol version 13.2 or later does not include this file. If you still want to use it, you can either copy it from a former Jmol version or get it from the Jmol repository.
Jmol Applet JAR files
Regardless of which JavaScript library is used, Jmol-JSO or Jmol.js, there are four different ways to implement Jmol as an applet:
- Monolithic unsigned applet You can include
 JmolApplet.jar on your web site and just use that. If using Jmol.js, the applet will only have access to files on your site; if using Jmol-JSO, the applet will have access to files on your site as well as files at public databases.
JmolApplet.jar on your web site and just use that. If using Jmol.js, the applet will only have access to files on your site; if using Jmol-JSO, the applet will have access to files on your site as well as files at public databases.
- Modular unsigned applet You can include
 JmolApplet0.jar and related files (54
JmolApplet0.jar and related files (54  jar files total) on your site. Although a large number of files, the advantage of the modular applet is that only those parts of Jmol that are necessary are downloaded by the user's browser initially, and additional components are transferred only as needed. This allows the applet to load much faster.
jar files total) on your site. Although a large number of files, the advantage of the modular applet is that only those parts of Jmol that are necessary are downloaded by the user's browser initially, and additional components are transferred only as needed. This allows the applet to load much faster.
- Monolithic signed applet
 JmolAppletSigned.jar is a large (2.5+ MB) single file that includes all of Jmol in the form of a signed applet. The signed applet has substantially more capability, including full cross-domain file access and the capability to transfer files to and from the user's hard drive.
JmolAppletSigned.jar is a large (2.5+ MB) single file that includes all of Jmol in the form of a signed applet. The signed applet has substantially more capability, including full cross-domain file access and the capability to transfer files to and from the user's hard drive.
- Modular signed applet
 JmolAppletSigned0.jar and related files, like JmolApplet0.jar, allows modular as-needed transfer of files from your site to the user's browser. This is the preferred method of using the signed applet.
JmolAppletSigned0.jar and related files, like JmolApplet0.jar, allows modular as-needed transfer of files from your site to the user's browser. This is the preferred method of using the signed applet.
Giving the Jmol applet more memory to work with
Note: This can now be done using Jmol.js (jmolSetMemoryMb(nMb)) or Jmol-JSO (Info.memoryLimit = nMb).
Memory available to Jmol is determined by Java. You can check the current memory allocation by opening the pop-up menu in the applet, and the last submenu "About Jmol".
Less or more memory can be allocated by using this parameter:
-Xmx###M
where the ### must be substituted by a number in megabytes (hence the "M" after it). However, the amount of memory that can be allocated is limited by your existing RAM. For example, on a system with 512 MB RAM, a maximum of 256 MB is allowed for Java.
Examples:
- -Xmx512M will give 512 Mb maximum memory available.
- -Xmx1024M will give 1024 Mb maximum memory available.
The place to set this parameter depends upon your system:
Windows
- Control Panel > Java icon (opens the Java Control Panel) > "Java" tab > "Applet run-time configuration" > "Show" button > on row "JRE" (latest version if there are several), click on the textbox under "Parameters" and type: -Xmx###M
Macintosh
- OS 10.5 (Leopard): Double click on JavaPreferences.app (in /Applications/Utilities). In the 'General' tab, select the first version of Java that is in the list. This will activate the 'Options' button: click it. A box for 'Applet Runtime Parameters' will open, where you can enter the -Xmx###M parameter.
- OS 10.4 (Tiger): Possibly look under /Applications/Utilities/Java/J2SE 5.0/Java Preferences, then look for the place to put the -Xmx###M
- Or look in the Java Control Panel.
Note that this is a local setting, so it will only affect that computer. You cannot expect that visitors of your web pages will have the same Java memory configuration as you, so be careful when using this feature.
Technical details:
- -Xmx sets the maximum memory heap size.
- -Xms sets the minimum memory heap size.
See also the procedure for the application.
Forcing the Jmol applet to open in a certain language
The Jmol applet includes several languages (technically called localizations) for the user interface (pop-up menu). The applet will open by default with the interface using the language of your operating system (ref.). However, if you need to change this:
- Starting with Jmol 11.1.30, the language in the applet can be switched anytime, from the pop-up menu, "Language" item near the bottom.
- Or, using the scripting language:
language = "de"(you must use the keyword "language" and the two-letter language code: ca, cs, de, en, es, et, fr, nl, pt, tr, etc.).
Testing different applet versions
(This is an advanced feature)
You can force the use of a certain version of Jmol applet in any web page (even if that page is not yours). The way to do so is:
On the URL line of the browser, add
?JMOLJAR=
followed by the url of the ![]() JmolApplet0.jar or
JmolApplet0.jar or ![]() JmolAppletSigned0.jar file your intend to use.
If there is already a
JmolAppletSigned0.jar file your intend to use.
If there is already a ? in the URL, then use &JMOLJAR=, to preserve the logic of a url.
There will be a couple of warnings that you will have to accept, due to the potential security risk of replacing an applet with another coming from a totaly unrelated server. There may also be Java security issues still preventing the applet from loading files, unless you specify the signed applet.
For example:
http://www.rcsb.org/pdb/explore/jmol.do?structureId=1CRN&bionumber=1&JMOLJAR=http://chemapps.stolaf.edu/jmol/docs/examples-12/JmolAppletSigned0.jar
lets you use Bob's latest version of Jmol at the RCSB site (which is currently using 12.2, signed). Try it.
If you just want a change to the signed version of the same applet, this shortcut notation may be used (Jmol.js in 12.2.RC8 or later):
?JMOLJAR=SIGNED
(But note that the website author may have omitted the signed applet files in the server, so the applet will fail to load.)
Jmol in Wikis
The applet can be embedded in HTML (or XHTML) pages (examples are listed on Websites Using Jmol), but also, with some extra technologies, in Wiki pages (see Jmol Processor).
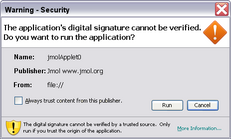
Unsigned vs. Signed Applet & Data Access
The signed applet can load a molecular model from any http or ftp URL. However, the user will be asked to trust the signed applet (figure at right). This could be problematic since the publishing authority (jmol.org) "cannot be verified", and the security certificate was issued by a group that is "not trusted" (jmol.org). This is because it would be costly and cumbersome for each version of the applet to be signed by a trusted (commercial) authority.
The unsigned applet displays the molecule without asking the user to trust it. However, it is limited (for security reasons) to loading molecular models from the same http domain (server) that downloads the applet itself. FirstGlance in Jmol uses the unsigned applet by default, but the signed applet will be used when that "Advanced Option" is checked. Thus you can easily see both applets in action. Methods for accessing data files from arbitrary servers are described in some detail at PDB Data File Access Methods.
If you are not running the applet from any server, i.e., you are using a web page residing in a local disk, further security restrictions are applied by Java: molecular models can only be located in the same folder/directory as the applet files or below it; read Jmol Applet Deployment Local.
Tools for Authoring Jmol Web Pages
Jmol.php: easy Jmol insertion into a web page
You can insert a Jmol applet into a web page with nothing more than a simple <script> or <a> tag.
You don't have to install any Jmol files in your computer or web server.
You don't even need a file with the 3D model; it will be automatically obtained from an appropriate web server.
Note that this way of using Jmol is easily compatible with any environment where you do not control the full source code, like forums, blogs, wikis, content management systems, e-learning environments... In fact, you can email the address and the recipient will see the Jmol applet by clicking on the received link.
See Jmol_PHP for details.
Proteopedia's Scene Authoring Tools
The easiest way to create, and share, a web page containing customized molecular scenes in Jmol, is to build the page in Proteopedia.Org. Proteopedia's Molecular Scene-Authoring Tools free you from learning any Jmol commands, HTML, or javascript. You will only need to learn a tiny bit of wikitext markup, for which convenience buttons and help are provided. On the other hand, if you already know some Jmol commands, you may use them in creating your scene (via the Jmol console). Regardless of how your scene is created, it will be saved as a Jmol state script. You may upload atomic coordinate files, or use published PDB files. Proteopedia automatically saves a frozen copy of each PDB file that you use, to protect your scripts from breaking e.g. after remediation of files by the Protein Data Bank. You may created pages that are protected from editing by others.
Jmol's Export to Web
If you are not comfortable writing html/xhtml and javascript code, or would just like to quickly put together some simple pages, then look at the export to web function of Jmol (available in version 11.4 and above). This allows you to export all the necessary files to make simple pages that have a number of live Jmol displays or a single Jmol display that can be changed by clicking buttons. You select the views you want the user to see by setting them up in Jmol. You then add any desired text or comments to the .html file created by the export function in your favorite web editor (Some open source suggestions: SeaMonkey is simple and works very well, Kompozer is more sophisticated and works pretty well, Amaya is the most sophisticated but is harder to use). Instructions for using the export function are accessible in the export to web dialog within the Jmol application. A more detailed tutorial and some older packages are available at Jmol Web Page Maker.
Saving Files from the Applet
Although this isn't obvious, the molecule that is being shown in the applet can be saved to local disk (even from the unsigned applet) unless the model was loaded inline, in which case this technique fails. To do so:
- Open the applet's menu (right-click, Ctrl+click, or click on bottom-right "Jmol" logo).
- Click on the last entry (
About...). - On the submenu that appears, the first entry is the name of the first model loaded. If it says "string" then the model was loaded inline and continuing will result in a "404" resource not found error. If the entry is a file name, then you can continue. If more than one file has been loaded, there will be a line for each on this submenu. Click on the one that you want to open or save. This will bring up another layer of menu options
- Click on the last entry (
View+ name of file) - Depending on your browser's configuration, the file will be opened in the associated program ("helper application") or you will get an "Open or Save" dialog; choose Save and location in your disk.
- In old Jmol versions:
- Open the applet's menu (right-click, Ctrl+click, or click on bottom-right "Jmol" logo).
- Click on topmost entry (name of molecule).
- On the submenu that appears, click on bottommost entry (name of model).
- Depending on your browser's configuration, the file will be opened in the associated program ("helper application") or you will get an "Open or Save" dialog; choose Save and location in your disk.
- Warning: At least in Windows, modern browsers may detect this action as opening a pop-up window, so it's likely that the pop-up blocker will prevent you from seeing the "Open or Save" dialog. Pay attention to messages and options in your browser to allow pop-ups for the current page. (Such messages often appear as yellow bars at the top of the page, warnings in a toolbar, or hints in the status line at the bottom of the browser window.)
- Note for Chime users: If you have MDL Chime installed, the molecule will be opened in a browser window using Chime. You just have to use Chime's menu (File > Save molecule as) to save the file to disk.
If the page uses the signed applet, there is a more intuitive and fully functional File submenu near the bottom of the popup menu.
In addition to saving the full model, a part of the molecule can be extracted and saved into MOL format. This is not so straightforward as the former, and is described in the "interactive scripting documentation" (see Jmol home page).
It is also possible to save the state of the molecule (rendering style, coloring, visibility, orientation, etc.). See save state and write state in the Jmol scripting documentation. This gives similar functionality to what can be done in Chime or Rasmol, although due to Java security restrictions, saving to a file is not available for the unsigned applet. Instead, you get the data in the console, from where you can copy to a text editor and then save from that.
Setting up a Jmol site in The Cloud, in an AWS S3 bucket
Oct 2013: This section describes how to set up a simple Jmol demonstration site in an S3 bucket. In passing it also confirms a working layout of JmolAppletxxx files, html files, model files as described above. Note this is purely for the Java version of Jmol, not JSmol.
AWS S3 (Amazon Web Services, Simple Storage Service) is ideal for setting up a simple demonstration Jmol site with embedded applets.
S3 provides reliable, cheap cloud storage, with a default web access service. Costs are negligible for a small site: http://aws.amazon.com/s3/pricing/ Also you get 12 months useful access free with the AWS Free Usage Tier. This gets around the many headaches of using web servers on institutional computers.
Example of S3 bucket used as Proteomics data archive, for academic paper Supplementary Information: http://mouse-brain-ptmome.s3.amazonaws.com/list.html
Example of S3 bucket used as a Jmol demo: http://ptmjmol.s3.amazonaws.com/section1/trace.html
Example of S3 bucket used as an image gallery: http://ptmgallery.s3.amazonaws.com/list.html
You probably already have an Amazon account for book purchase. If not, https://www.amazon.com and navigate to New Account. Then go to http://aws.amazon.com/console/ and sign in. Go to the S3 Console by clicking on "S3 Scalable Storage In The Cloud", or select S3 from the Services menu at the top left.
Create a bucket. Bucket names are unique across all the AWS data centres, so obvious ones like "test" or "mybucket" will already be taken. The bucket will be accessible at the base address http://bucketname.s3.amazonaws.com It will also be accessible via http://s3.amazonaws.com/bucketname/
The S3 Console provides simple facilites to create buckets, and folders and subfolders within them, and upload/download files, and also to set file permissions and Mime file types, and other general filesystem admin (rename, delete, cut/copy/paste etc). There are other methods such as commandline tools, and Gui-based apps (see below) which allow more streamlined control for large numbers of files or repetitive tasks.
A layout which works for model files local in the bucket is ..
bucketname
Jmol.js
JmolApplet.js
JmolApplet.jar
JmolApplet0.jar
JmolApplet0_Console/Minimize/Popup/ReadersCifPdb/ReadersMolXyz/ReadersMore/ReadersQuantum/ReadersSimple.jar
JmolApplet0_ReadersXml/ReadersXtal/ShapeBio/ShapeSpecial/ShapeSurface/Smiles/Spectrum/Symmetry.jar
section1
mydemo.html
models
1GFL.pdb
4E50.pdb
Key script settings are ..
<html>
...
<script>
jmolInitialize("..", window.location.protocol=="file:");
...
jmolApplet(600, "load ../models/1GFL.pdb");
...
In the bucket, all files should generally have Public Access permissions. HTML files should have type text/html, images should have type image/png etc. If these do not set themselves automatically during your file upload, they can be corrected in the S3 Properties - Permissions/Metadata gui. Note it is easy to lose these settings when re-uploading a new version of a file.
Software revisions were:
Jmol 13.0.18 (installed in S3 bucket as per above .js and .jar files) Browsers used in testing: Google Chrome 30.0, Firefox 24.0, Safari 5.1, Opera 12.16 Most development on Mac OS X 10.6.8, java 1.6.0_51. Tested on 10.8.5, Windows 7.
Some free tools to make S3 bucket management easier:
http://aws.amazon.com/developertools/739 ... a command-line tool http://www.timkay.com/aws/ ... another link to the above http://s3browser.com/ ... a good Windows S3 bucket manager with nice Gui.
Contributors
AngelHerraez, GregE, EricMartz, Hansonrstolaf, Igor, NicolasVervelle, Gutow, Ted, Pimpim, Cudo29, Green